Green Spring's Fermentation-Based CoQ10 Powder Overcomes Bioavailability Limits
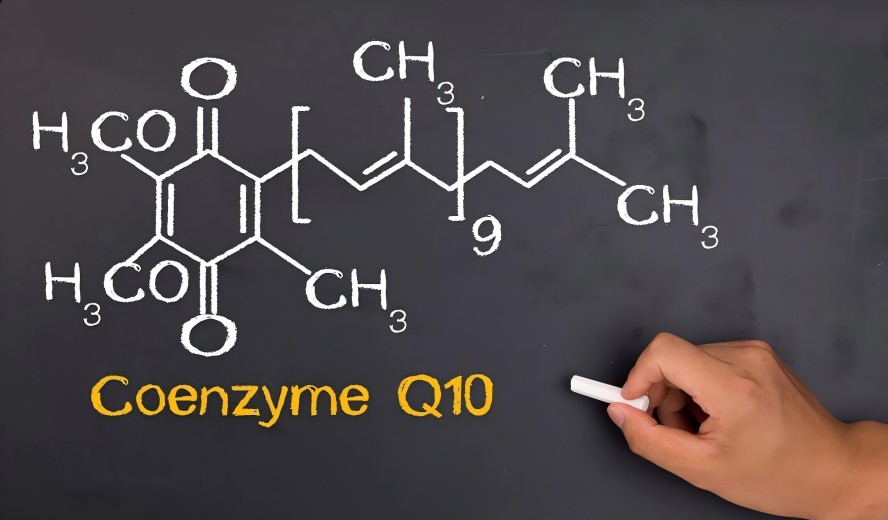
Coenzyme Q10, with its exceptional physiological functions, finds extensive applications in cardiovascular health, fatigue resistance, and skincare. However, as a large-molecule lipophilic substance, its inherently low absorption rate poses a core bottleneck limiting product efficacy. Furthermore, inconsistent raw material sources and varying quality standards in the market present challenges to the stability and safety of end-user brands.
To overcome this bioavailability barrier, the industry consensus is to produce CoQ10 raw materials through microbial fermentation. This method yields CoQ10 structurally identical to its natural form in the human body, avoiding ineffective isomers potentially generated by chemical synthesis. This ensures high biological activity and absorption potential from the source.
Green Spring Technology has deeply cultivated fermentation technology. Our CoQ10 raw material goes beyond mere “fermentation production”; through end-to-end innovation, we elevate quality and efficacy to new heights:
★ High Bioavailability
Using proprietary strains and precision fermentation processes, we guarantee 100% all-trans, high-potency CoQ10. Its molecular structure is precisely recognized and utilized by the human body, providing fundamental assurance for your product efficacy.
★ Full Traceability
We maintain a comprehensive traceability system from strain repository → fermentation → purification → finished product. Each batch carries a complete “identity profile,” guaranteeing purity, stability, and zero contamination—safeguarding your supply chain security and brand reputation.
Choosing Green Spring Technology's fermented CoQ10 raw material means acquiring more than just an ingredient—it grants you formidable market competitiveness:
◆ Exceptional Product Performance
With this highly bioavailable raw material, your end products can more effectively deliver to target sites, demonstrating superior functional efficacy and earning consumer acclaim.
◆ Solid Credentials
Our “full traceability” system provides transparent supply chain narratives and rigorous quality data—powerful tools for projecting a “safe, professional, and trustworthy” brand image to the market.
◆ Long-Term Viability
We guarantee high consistency between raw material batches, helping your products achieve stable quality and sustained market longevity.
1 From Source Strains to Final Ingredients: Defining New Quality Standards for Coenzyme Q10 with Cutting-Edge Biotechnology
In today's health-conscious world, the efficacy of Coenzyme Q10 is widely recognized, yet its absorption efficiency (bioavailability) remains the core bottleneck limiting product effectiveness. Green Spring Technology understands that overcoming this bottleneck begins at the source. We have abandoned chemically synthesized methods with demanding conditions and complex product isomers, steadfastly choosing microbial fermentation—the method offering optimal compatibility with the human body.
1.1 Why is fermentation the cornerstone of high bioavailability?
Unlike extraction from animal organs or chemical synthesis, Green Spring Technology employs highly efficient strains from the Rhodobacillaceae family, such as Rhodobacter sphaeroides, for fermentation. These microorganisms naturally synthesize all-trans CoQ10 identical to the human form, ensuring exceptional biological activity and recognition. This enables direct, efficient absorption and utilization by the body, fundamentally resolving the challenge of low bioavailability.
1.2 How does microbial strain engineering ensure purity and activity?
Wild strains have limited yields, so we precisely modify them using advanced metabolic regulation breeding techniques:
■ Precision Breeding to Strengthen Pathways
By selecting carotenoid-deficient mutant strains, we successfully blocked the competitive metabolic pathway for β-carotene synthesis. This redirects precursor substances more intensively toward CoQ10 synthesis, significantly boosting its content and purity. Both literature and our practice confirm this method increases CoQ10 yield by over 250%.
■ Unlocking Potential Through Inhibition Removal
We screened mutant strains resistant to metabolic inhibitors like vitamin K3 and ethionine. These strains successfully bypass feedback inhibition in the synthesis pathway, effectively “turning on the metabolic switch.” This fully unleashes CoQ10 synthesis capacity, ensuring high product potency.

1.3 Full Traceability: What Does “From DNA to Finished Product” Entail?
“Full traceability” extends beyond process documentation to represent an ultimate pursuit of source consistency and quality stability.
▲ Traceable Genetic Origins
Our patented strain repository serves as the starting point for full traceability. Every production batch originates from these rigorously screened and genetically verified pure strains, guaranteeing consistency and reliability of starting materials at the DNA level.
▲ Controlled Production Process
From strain activation and fermentation to isolation and purification, the entire process operates under a stringent quality control system. This ensures contamination-free, mutation-free production with product purity consistently maintained above 99%.
▲ Verifiable Quality Outcomes
Each batch of raw materials is accompanied by a comprehensive Certificate of Analysis (COA), including safety metrics such as heavy metals and solvent residues, along with High-Performance Liquid Chromatography (HPLC) profiles, guaranteeing high purity and bioactivity.
Through its innovative combination of “cutting-edge strains + precision fermentation + full-chain quality control,” Green Spring Technology has not only overcome the bioavailability bottleneck of Coenzyme Q10 but also provides customers with unprecedented quality assurance through its fully traceable system. We deliver not just raw materials, but solutions that ensure the safety and efficacy of your end products.
2 Green Spring Technology Ensures High Activity and Traceability of Coenzyme Q10
At Green Spring Technology, we precisely control the entire fermentation process to ensure every batch of CoQ10 raw material possesses exceptional biological activity and traceability.
2.1 Precision Culture Medium Design: Foundation for High Activity
We utilize scientifically balanced carbon and nitrogen sources (e.g., glucose-sucrose blends, yeast extract-corn steep liquor combinations) to continuously supply essential precursors, ensuring correct molecular structure. Precisely added trace elements like Mg²⁺ and Fe²⁺ serve as key enzyme cofactors, efficiently driving the CoQ10 synthesis pathway.
2.2 Dynamic Environmental Control: Ensuring Batch Stability and Consistency
We standardize critical parameters including temperature (28-30°C), pH (6.5-7.0), and dissolved oxygen to create an optimal growth environment for the microbial culture. This not only guarantees high reproducibility of metabolic pathways but also enables process-level control and traceability of quality.
2.3 Intelligent Precursor Supplementation: Targeted Enhancement of Yield and Activity
During critical fermentation stages, precise addition of side-chain precursors (e.g., ubiquinol) and quinone ring precursors (e.g., p-hydroxybenzoic acid) effectively strengthens the target synthesis pathway. This strategy boosts yield by over 90% while ensuring the integrity and high activity of CoQ10 molecules.

3 Green Spring Achieves Full Traceability Throughout the Quality Process
At Green Spring Technologies, we regard separation and purification as the “final mile” in safeguarding CoQ10's biological activity. Improper control at this stage can easily render the high-activity molecules meticulously cultivated during fermentation useless due to oxidation or structural damage. Therefore, we have established a purification process system centered on the core principles of “gentle, precise, and verifiable,” ensuring the highest product quality and stable reliability.
3.1 Gentle Targeted Extraction: Safeguarding Molecular Integrity at the Source
To avoid the risk of methoxy group substitution and the formation of ineffective derivatives associated with traditional alcohol-alkali saponification, Green Spring Technology prioritizes advanced low-temperature hydrocarbon solvent targeted extraction technology.
■ Mitigating Structural Risks
Through precise control of process parameters, we completely eliminate potential molecular structural substitution side reactions that may occur under strong alkaline, high-temperature conditions. This ensures every CoQ10 molecule in the final product possesses the fully bioactive natural all-trans structure. This is the molecular foundation for guaranteeing high bioavailability.
■ Comprehensive Antioxidant Protection
From the outset of purification, we incorporate food-grade antioxidants (such as pyrogallic acid) and implement oxygen-free operations throughout subsequent concentration and chromatography steps. This creates an oxygen-free purification environment, effectively preventing CoQ10 inactivation due to oxidation.
3.2 Precise Chromatographic Purification
Following crude extraction, we employ high-performance silica adsorption column chromatography as a critical refinement step.
▲ Precise Separation
By optimizing the polarity and gradient of elution solvents (e.g., diethyl ether-petroleum ether mixtures), we achieve molecular sieve-like precision in separating target CoQ10 from other lipophilic impurities and byproducts.
▲ Data-Driven Control
Critical parameters—including elution curves and collection intervals—are standardized and monitored digitally. This ensures highly consistent purification outcomes across production batches and provides process-level traceability for each product's purity.
3.3 Crystallization and Quality Control: The Endpoint and Starting Point of Quality Traceability
Finally, through vacuum distillation and crystallization with anhydrous ethanol, we obtain high-purity orange-yellow CoQ10 crystals.
◆ Crystallization Ensures Activity
Mild crystallization conditions further guarantee stable crystal morphology and enhanced bioavailability.
◆ Comprehensive Quality Profile
Simultaneously, a comprehensive “Purification History” is generated. This document meticulously records data including solvent types used, residual levels, and key impurity control thresholds for the batch. Combined with upstream strain and fermentation data, this history forms a complete traceability system for CoQ10 raw materials—from gene to finished product-providing robust data support for customer product safety and regulatory compliance.

4 Precision Analysis Locks in High Activity; Data-Driven Closed-Loop Ensures Traceability
At Green Spring Technologies, we believe exceptional quality stems not only from precision manufacturing but also from rigorous validation. Our analytical identification system is the scientific foundation that delivers on our promise to overcome bioavailability bottlenecks and enables full traceability to the final product. We employ multidimensional analytical techniques to establish indisputable quality profiles for every batch of CoQ10 raw material.
4.1 Beyond Purity: Focusing on Precise Structural and Activity Verification
We understand that meeting content standards alone does not guarantee high bioavailability. Therefore, our analytical focus centers on confirming the molecular structural integrity and high purity of CoQ10.
▲ UV Spectrophotometry
While recognizing its convenience, we acknowledge potential errors from impurity interference in complex samples. At Green Spring Technology, this method serves solely as an auxiliary tool for rapid screening and process monitoring, never as the final quality criterion.
▲ High-Performance Liquid Chromatography (HPLC)
We mandate HPLC as the core basis for product release. This method precisely separates CoQ10 from any potential impurities, providing accurate content and purity data. By comparing retention times and UV spectra between samples and reference standards, we ensure the structural integrity of the product's main component.
4.2 Activity Validation: Exclusive “All-Trans” Configuration Confirmation
To scientifically validate the product's high biological activity, we implement a rigorous identification protocol:
● Reduced State Validation Test
We conduct proprietary chemical verification by exploiting CoQ10's characteristic peak disappearance in its reduced state (upon addition of sodium borohydride). This confirmatory test establishes it as physiologically active oxidized CoQ10, not other interfering substances.
● Pursuing Precise Configuration Analysis
To ensure the product exhibits 100% all-trans active configuration (critical for high bioavailability), Green Spring Technology has implemented advanced analytical techniques (such as chiral chromatography or LC-MS) to absolutely confirm the product's stereochemical structure, fundamentally eliminating the presence of inactive isomers.
4.3 Data Closure: Providing Final Verification for “Full Traceability”
All analytical identification processes culminate in a comprehensive Certificate of Analysis (COA).
◆ From Data to Credibility
This report delivers more than just a “Purity ≥99%” figure. It includes HPLC chromatograms, UV absorption spectra, moisture and solvent residue levels, heavy metals, and other safety metrics. It serves as a complete “product identity document.”
◆ Complete Quality Traceability
Each batch's CoA is tightly linked to its microbial strain batch number, fermentation tank number, and purification batch. This means customers receive not just raw materials, but a fully transparent data chain traceable back to the production source. This truly achieves quality confidence transfer from “manufacturing” to “application,” providing the most robust scientific support for your product safety and market claims.
5 Green Spring Technology's CoQ10 Ingredients Empower Your Product Formulation Innovation
Amid the global expansion of the CoQ10 market, technological breakthroughs and quality upgrades have become the core drivers of industry development. As China's first enterprise to achieve large-scale, precision production of fermented CoQ10, Green Spring Technology leverages its independently developed end-to-end technology platform—spanning strain construction, fermentation regulation, and separation/purification—to successfully achieve stable mass production of high bioavailability CoQ10 powder, setting a new quality benchmark for the industry. Discover Our Comprehensive Solution for Compliant and Stable Coenzyme Q10 Powder.
By establishing a comprehensive traceability system covering the entire production process, Green Spring ensures every batch of CoQ10 raw material delivers consistent high bioactivity and reliable quality performance, completely resolving the industry pain points of low activity and batch inconsistency in traditional raw materials.
We remain focused on optimizing microbial strain performance and upgrading production processes, achieving continuous breakthroughs in synthetic biology and intelligent fermentation. This enables us to provide downstream enterprises with superior, more reliable core raw materials.
For more product information or to request samples, please contact us at helen@greenspringbio.com or WhatsApp: +86 13649243917.
References:
[1] WU Zufang,et al. Progress of functional studies on coenzyme Q10[J]. Journal of Ningbo University(Science and Technology),2001 , 14(2):85-88.
[2] Yuan Yi. Extraction and purification of coenzyme Q10 from pig heart[J]. Journal of Anhui Agricultural University, 1997,24(2):200-203. [3] Zhao Meifa.
[3] Zhao Meifa. Coenzyme Q10 is an important prodrug[J]. Fine and Specialty Chemicals 2001, (22):35-40.
(22):35-40.
[4] GU Jue-Fen ,CHEN Guang-Ming ,LI Shu-Zhen. Synthesis of coenzyme Q10 by microbial transformation [J]. Pharmaceutical Progress,2001,25(6):339-343.
[5] MEGANATHAN R. Ubiquinone biosynthesis in microorganisms [J]. FEMS Microbiol Lett, 2001,203:l3l-l39.
[6] Liu K-S,Wu W-F,Han S-Q,et al. Selection and development of high-yielding strains of coenzyme Q10[J]. Journal of Shenyang Agricultural University,2005,36(1):89-92.
[7] ZHANG Yanjing , YUAN Qipeng , LIANG Hao. Study on the fermentation conditions of Coenzyme Q10-producing yeast[J]. Microbiology Bulletin,2003,30(2):65-69.
[8] OLSON E O, RUDNEY H. Biosynthesis of ubiquinone [J]. vitam Horm, 1983 ,40:1-43.
[9] YOSHIDA H, KOTANI Y, OcHIAI K, et al. production of u- biquinone-10 using bacteria[J]. J Gen Appl Microbiol,1998,44: 19-26.
[10] TEIZIL U. production of ubiquinone and bacteriochlorophy Ⅱ a by Rhodobacter sphaeroides and Rhodobacter sulfidophilus [J]. J Fermen Bioeng, 1993 ,76: 191-194.
-
Prev
Reduced CoQ10 Raw Material: Powering Next-Gen Supplement Innovation
-
Next
Green Spring Technology's 98% Coenzyme Q10 Powder for Heart Health Support


 English
English French
French Spanish
Spanish Russian
Russian Korean
Korean Japanese
Japanese




